Ampure Beads Size Selection Chart
SPRI Based Size Selection Introduction SPRIselect is a SPRI-based chemistry that speeds and simplifies nucleic acid size selection for fragment library preparation for Next Generation sequencing. Use a 1X ratio of beads to library to remove fragments shorter than 200 bp.
 Cleanngs Spri Beads From Bulldog Bio Biocompare
Cleanngs Spri Beads From Bulldog Bio Biocompare
Find high-quality picture Ampure beads size selection chart
Add 40ul of beads to this supernatant and wait 15min pelletwash the beads by EtOH and elute the DNA in x ul of Tris.. Additionally the DNA concentration of the SMRTbell library to be size selected must be 0510 ngµL. AxyPrep MAG PCR Clean-Up Kit 5 mL 227 preps MAG-PCR-CL-5. AxyPrep AMPure XP MinElute.
Are functionally similar to AMPure XP beads. Tunable fragment size selection to suit specific applications. The result is a more purified PCR product.
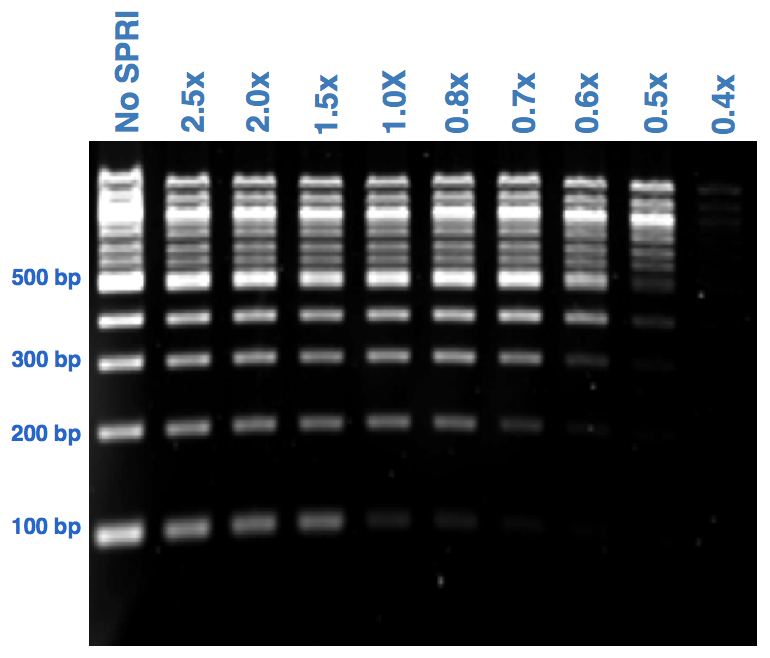
Different bead ratios can be used depending on the protocol and desired size selection. 06x-1x dual size selection. If you perform the QC check and your sample contains Adaptor dimer 127 peak or excess primers 70-80 it is recommended to use gel or Pippin Prep for size selection.
Beads are used for DNA purification andor size selection of DNA fragments based on the ratio of beadssample. Predictable consistent size selection between runs and reagent lots. Prepare your DNA in 100ul Tris or Elution buffer from BioO etc add 60ul of bead wait 15min and transfer 160ul supernatant to a new well.
To the purified PCR reaction 25 μl add 325 μl 13X of resuspended AMPure XP beads and mix well on a vortex mixer or by pipetting up and down at least 10 times. You can use our proprietary SPRI paramagnetic bead-based chemistry to remove contaminants dNTPs salts primers primer dimers throughout your NGS workflows. Beads are used for DNA purification andor size selection of DNA fragments based on the ratio of beadssample.
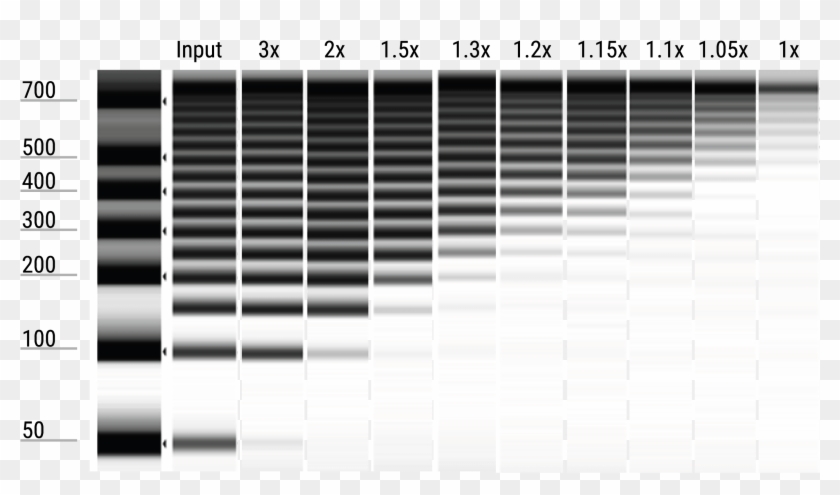
SPRI enables our chemistries to deliver high-performance isolation. CHART COLOR BEADSAMPLE RATIO PINK InputReference RED 18X BLUE 09X GREEN 07X AQUA 06X AMPure XP. Solid Phase Reversible Immobilization SPRI technology uses paramagnetic beads to selectively bind nucleic acids by type and size.
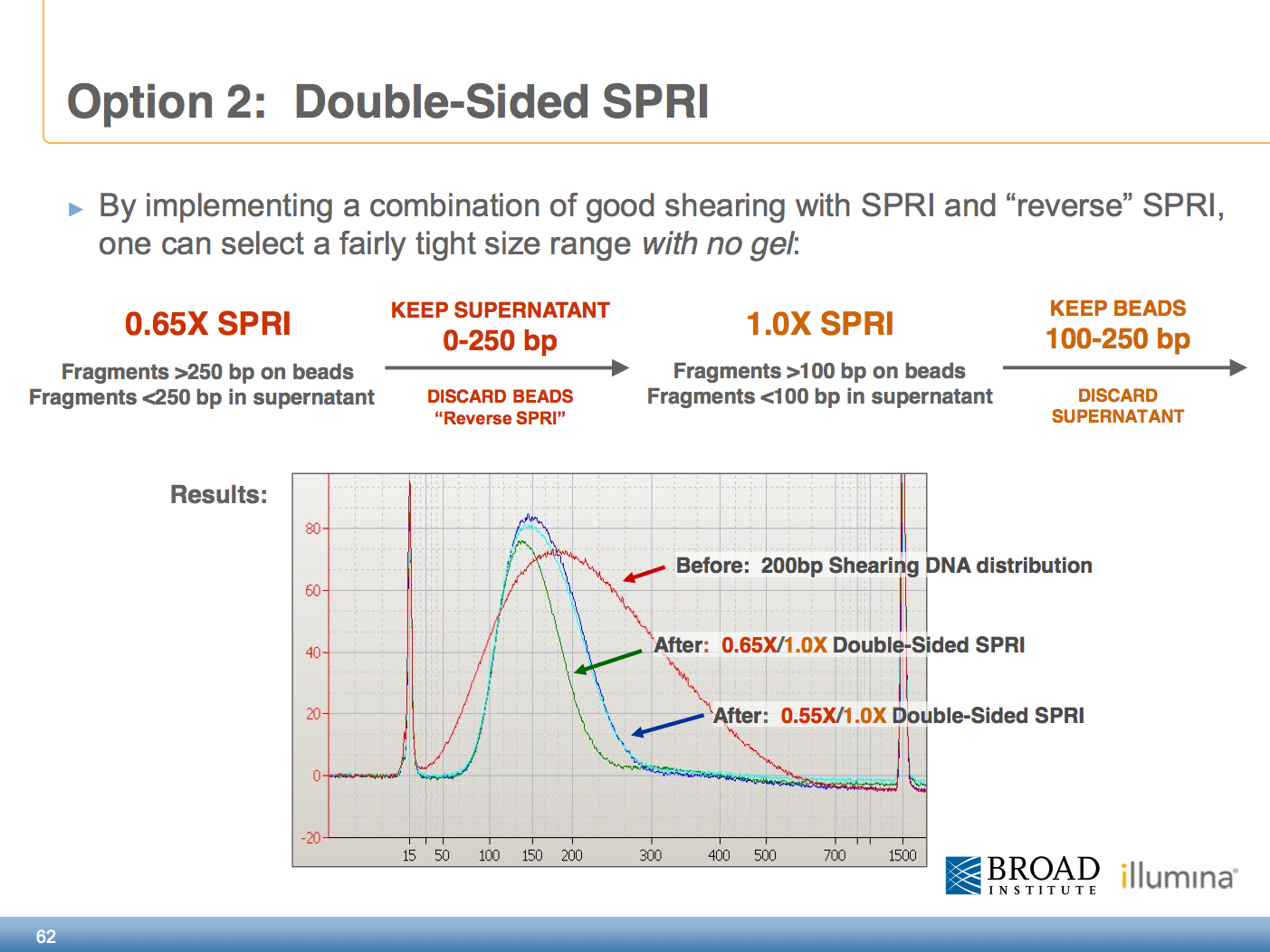
Example in the procedure I follow we are interested in selections fragment size between 250-450 bps. Use a 06X ratio of beads to library to remove fragments up to 300-350 bp. IPB Illumina Purification Beads Illumina DNA PCR-Free Prep Tagmentation.
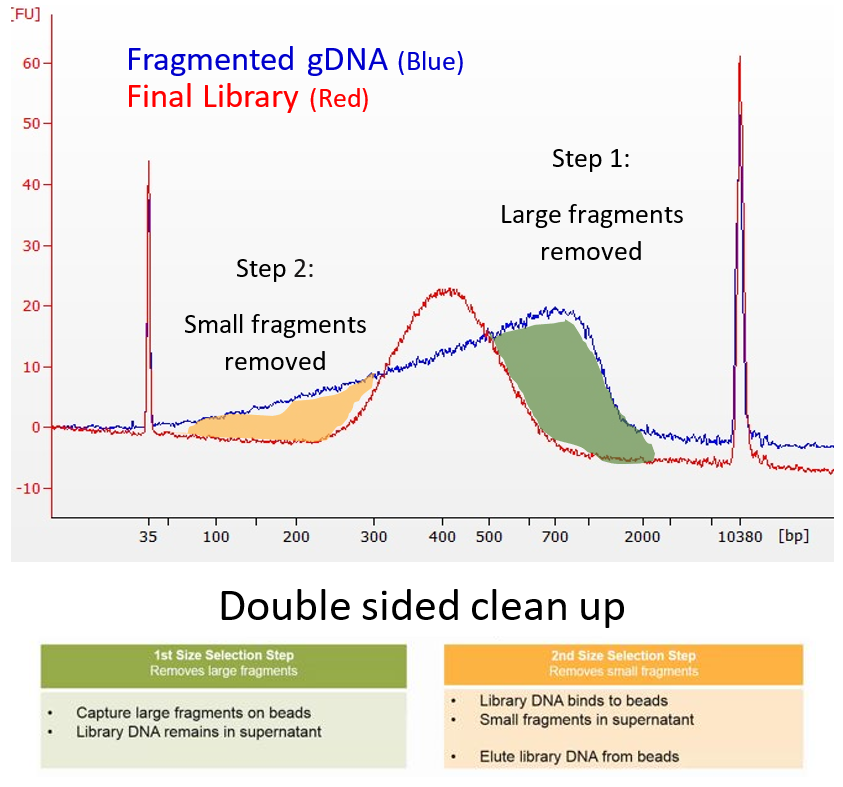
Agilent TapeStation traces of sheared gDNA purified using Ampure XP Reagent O and Reagent K. Size selection using AMPure XP Beads does not remove small fragments. Incubate for 5 minutes at room temperature.
In this process size selection is required to produce a uniform distribution of fragments around an average size. The first article weve found that describes this selection is referenced below 3. Predictable and consistent size selection.
Promega Beads Promega. Use of higher concentrations. Quality Control Check and Size Selection using AMPure XP Beads NEBNext Small RNA Library Prep Set for Illumina E7300 E7580 E7560 E7330 6C.
AMPure XP for PCR Purification Cleanup and Size Selection. Agencourt AMPure XP purified products can be used in the following applications. Agencourt AMPure XP utilizes an optimized buffer to selectively bind DNA fragments 100 and larger to paramagnetic beads.
Magnetic Beads Kits Kit Size Cat. Size-selection using Diluted AMPure PB Beads For effective size-selection using AMPure PB beads accurate pipetting is necessary. To the purified PCR reaction 25 μl add 325 μl 13X of resuspended AMPure XP beads and mix well on a vortex mixer or by pipetting up and down at least 10 times.
AMPure XP- Gold Standard for bead based Next-Generation Sequencing NGS clean-up. Size Select the small RNA library using AMPure XP beads after using column purification. High recovery of amplicons 100 bp.
Compatible with manual and automated processing. Recommended Conditions for Dual Bead-based Size Selection. Incubate for 5 minutes at room temperature.
CHART COLOR BeadSample Ratio PINK InputReference RED 18X BLUE 09X GREEN 07X AQUA 06X Figure 2. Purify DNA fragments of the desired size for library preparation using our proprietary SPRI paramagnetic bead-based chemistry which provides. Agencourt AMPure and Qiagen MinElute.
Throughput optimized DNA size selection suitable for various next generation sequencing. Size Select the small RNA library using AMPure XP beads after using column purification. For AMPure XP Bead-based Size Selection expect size distributions in the range of 230270 for 100 reads and 310370 for 200 reads.
Consult the appropriate library prep user guide for further details on the ratios. Clean-ups were performed using 18x red 09x blue 07x green and 06x aqua reagent to sample ratios as shown. Purify the PCR amplified cDNA construct 100 μl using a QIAQuick PCR Purification Kit.
SPRIselect for Size Selection. Excess primers nucleotides salts and enzymes can be removed using a simple washing procedure. Adding 35ul of AMPure XP beads it allows to catch the fragment size.
Kapa Pure Beads Kapa Biosystems. Known for use in our Agencourt AMPure XP which uses paramagnetic beads to selectively bind nucleic acids by type and size. This bead is d at room temperature.
Mag-Bind Omega Biotek. Bead size selection is only recommended for samples showing no primer dimer and no adaptor dimer on Bioanalyzer. Used in a variety of NGS library prep chemistries.
Compatibility with manual and automated processing. Since this publication on May 7th 2014 there are several more commercial Ampure-like size selection beads on the market. Add 40ul of beads to this supernatant and wait 15min pelletwash the beads by EtOH and elute the DNA in x ul of Tris.
 Dna Size Selection Chemistry Ampure Bead Size Selection Clipart 3770951 Pikpng
Dna Size Selection Chemistry Ampure Bead Size Selection Clipart 3770951 Pikpng
 Size Selection Using Ampure Xp Beads Neb
Size Selection Using Ampure Xp Beads Neb
 Optimising Library Size Selection For Iclip2 Sample To Pronex Bead Scientific Diagram
Optimising Library Size Selection For Iclip2 Sample To Pronex Bead Scientific Diagram
Plos One Unlocking Short Read Sequencing For Metagenomics
 Kapa Pure Beads Roche Sequencing Solutions
Kapa Pure Beads Roche Sequencing Solutions
 Next Generation Sequencing Tips N Tricks Part 2 Diagnostech
Next Generation Sequencing Tips N Tricks Part 2 Diagnostech
 Schematic Workflow Of Benus The Benus Method Was Constructed Using Two Scientific Diagram
Schematic Workflow Of Benus The Benus Method Was Constructed Using Two Scientific Diagram
 Size Dependent Isolation Of Dna Fragments From Sheared Genomic Dna Via Scientific Diagram
Size Dependent Isolation Of Dna Fragments From Sheared Genomic Dna Via Scientific Diagram
 Size Dependent Isolation Of Dna Fragments From Sheared Genomic Dna Via Scientific Diagram
Size Dependent Isolation Of Dna Fragments From Sheared Genomic Dna Via Scientific Diagram
 Size Dependent Isolation Of Dna Fragments From Sheared Genomic Dna Via Scientific Diagram
Size Dependent Isolation Of Dna Fragments From Sheared Genomic Dna Via Scientific Diagram
 Dna Purification Magnetic Beads 5 Ml
Dna Purification Magnetic Beads 5 Ml
 Spri Beads Dna Rna Purification High Quality Low Cost Biodynami
Spri Beads Dna Rna Purification High Quality Low Cost Biodynami

 How Do Spri Beads Work Enseqlopedia
How Do Spri Beads Work Enseqlopedia
 How Do Spri Beads Work Enseqlopedia
How Do Spri Beads Work Enseqlopedia
 Size Selection Using Ampure Xp Beads Neb
Size Selection Using Ampure Xp Beads Neb
Highprep Pcr Beads As An Ampurexp Alternative Genomics Core At Nyu Cgsb
 Next Generation Sequencing Tips N Tricks Part 2 Diagnostech
Next Generation Sequencing Tips N Tricks Part 2 Diagnostech
Ls Beckmancoulter Jp Files Appli Note Gel Using Spriselect

 Optimising Library Size Selection For Iclip2 Sample To Pronex Bead Scientific Diagram
Optimising Library Size Selection For Iclip2 Sample To Pronex Bead Scientific Diagram
